Lecture Week 3 Fri 10/19#
import seaborn as sns
df = sns.load_dataset("penguins")
what is the average body mass for adelie penguins#
df
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Gentoo | Biscoe | NaN | NaN | NaN | NaN | NaN |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
344 rows × 7 columns
df[df.species == 'Adelie'].body_mass_g.mean()
3700.662251655629
df.species
0 Adelie
1 Adelie
2 Adelie
3 Adelie
4 Adelie
...
339 Gentoo
340 Gentoo
341 Gentoo
342 Gentoo
343 Gentoo
Name: species, Length: 344, dtype: object
df.species == 'Adelie'
0 True
1 True
2 True
3 True
4 True
...
339 False
340 False
341 False
342 False
343 False
Name: species, Length: 344, dtype: bool
df[df.species == 'Adelie']
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 147 | Adelie | Dream | 36.6 | 18.4 | 184.0 | 3475.0 | Female |
| 148 | Adelie | Dream | 36.0 | 17.8 | 195.0 | 3450.0 | Female |
| 149 | Adelie | Dream | 37.8 | 18.1 | 193.0 | 3750.0 | Male |
| 150 | Adelie | Dream | 36.0 | 17.1 | 187.0 | 3700.0 | Female |
| 151 | Adelie | Dream | 41.5 | 18.5 | 201.0 | 4000.0 | Male |
152 rows × 7 columns
df[df.species == 'Adelie'].body_mass_g
0 3750.0
1 3800.0
2 3250.0
3 NaN
4 3450.0
...
147 3475.0
148 3450.0
149 3750.0
150 3700.0
151 4000.0
Name: body_mass_g, Length: 152, dtype: float64
df[df.species == 'Adelie'].body_mass_g.mean()
3700.662251655629
df.groupby('sex')['body_mass_g'].mean()
sex
Female 3862.272727
Male 4545.684524
Name: body_mass_g, dtype: float64
df.describe()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| count | 342.000000 | 342.000000 | 342.000000 | 342.000000 |
| mean | 43.921930 | 17.151170 | 200.915205 | 4201.754386 |
| std | 5.459584 | 1.974793 | 14.061714 | 801.954536 |
| min | 32.100000 | 13.100000 | 172.000000 | 2700.000000 |
| 25% | 39.225000 | 15.600000 | 190.000000 | 3550.000000 |
| 50% | 44.450000 | 17.300000 | 197.000000 | 4050.000000 |
| 75% | 48.500000 | 18.700000 | 213.000000 | 4750.000000 |
| max | 59.600000 | 21.500000 | 231.000000 | 6300.000000 |
# loc : use row and column label
df.loc[0:2, ['species', 'island']]
| species | island | |
|---|---|---|
| 0 | Adelie | Torgersen |
| 1 | Adelie | Torgersen |
| 2 | Adelie | Torgersen |
# iloc : use row and column number
# follow python convention, end point not included
df.iloc[0:2, 0:3]
| species | island | bill_length_mm | |
|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 |
| 1 | Adelie | Torgersen | 39.5 |
df.iloc[0:2, [1,3,5]]
| island | bill_depth_mm | body_mass_g | |
|---|---|---|---|
| 0 | Torgersen | 18.7 | 3750.0 |
| 1 | Torgersen | 17.4 | 3800.0 |
Q: What is the bill length of the heaviest penguin in the dataset?#
df.body_mass_g
0 3750.0
1 3800.0
2 3250.0
3 NaN
4 3450.0
...
339 NaN
340 4850.0
341 5750.0
342 5200.0
343 5400.0
Name: body_mass_g, Length: 344, dtype: float64
df.body_mass_g.idxmax()
237
df.loc[df.body_mass_g.idxmax()]
species Gentoo
island Biscoe
bill_length_mm 49.2
bill_depth_mm 15.2
flipper_length_mm 221.0
body_mass_g 6300.0
sex Male
Name: 237, dtype: object
df.loc[df.body_mass_g.idxmax(),'bill_length_mm']
49.2
Q: How many penguins are on Dream island and have body mass greater than 4000 grams?#
df['island'] == 'Dream'
0 False
1 False
2 False
3 False
4 False
...
339 False
340 False
341 False
342 False
343 False
Name: island, Length: 344, dtype: bool
df['body_mass_g'] > 4000
0 False
1 False
2 False
3 False
4 False
...
339 False
340 True
341 True
342 True
343 True
Name: body_mass_g, Length: 344, dtype: bool
(df['island'] == 'Dream') & (df['body_mass_g'] > 4000)
0 False
1 False
2 False
3 False
4 False
...
339 False
340 False
341 False
342 False
343 False
Length: 344, dtype: bool
df[(df['island'] == 'Dream') & (df['body_mass_g'] > 4000)]
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 35 | Adelie | Dream | 39.2 | 21.1 | 196.0 | 4150.0 | Male |
| 39 | Adelie | Dream | 39.8 | 19.1 | 184.0 | 4650.0 | Male |
| 43 | Adelie | Dream | 44.1 | 19.7 | 196.0 | 4400.0 | Male |
| 45 | Adelie | Dream | 39.6 | 18.8 | 190.0 | 4600.0 | Male |
| 49 | Adelie | Dream | 42.3 | 21.2 | 191.0 | 4150.0 | Male |
| 91 | Adelie | Dream | 41.1 | 18.1 | 205.0 | 4300.0 | Male |
| 93 | Adelie | Dream | 39.6 | 18.1 | 186.0 | 4450.0 | Male |
| 95 | Adelie | Dream | 40.8 | 18.9 | 208.0 | 4300.0 | Male |
| 97 | Adelie | Dream | 40.3 | 18.5 | 196.0 | 4350.0 | Male |
| 99 | Adelie | Dream | 43.2 | 18.5 | 192.0 | 4100.0 | Male |
| 133 | Adelie | Dream | 37.5 | 18.5 | 199.0 | 4475.0 | Male |
| 139 | Adelie | Dream | 39.7 | 17.9 | 193.0 | 4250.0 | Male |
| 146 | Adelie | Dream | 39.2 | 18.6 | 190.0 | 4250.0 | Male |
| 160 | Chinstrap | Dream | 46.0 | 18.9 | 195.0 | 4150.0 | Female |
| 165 | Chinstrap | Dream | 52.0 | 18.1 | 201.0 | 4050.0 | Male |
| 167 | Chinstrap | Dream | 50.5 | 19.6 | 201.0 | 4050.0 | Male |
| 171 | Chinstrap | Dream | 49.2 | 18.2 | 195.0 | 4400.0 | Male |
| 177 | Chinstrap | Dream | 52.0 | 19.0 | 197.0 | 4150.0 | Male |
| 181 | Chinstrap | Dream | 52.8 | 20.0 | 205.0 | 4550.0 | Male |
| 183 | Chinstrap | Dream | 54.2 | 20.8 | 201.0 | 4300.0 | Male |
| 185 | Chinstrap | Dream | 51.0 | 18.8 | 203.0 | 4100.0 | Male |
| 189 | Chinstrap | Dream | 52.0 | 20.7 | 210.0 | 4800.0 | Male |
| 191 | Chinstrap | Dream | 53.5 | 19.9 | 205.0 | 4500.0 | Male |
| 197 | Chinstrap | Dream | 50.8 | 18.5 | 201.0 | 4450.0 | Male |
| 199 | Chinstrap | Dream | 49.0 | 19.6 | 212.0 | 4300.0 | Male |
| 205 | Chinstrap | Dream | 50.7 | 19.7 | 203.0 | 4050.0 | Male |
| 209 | Chinstrap | Dream | 49.3 | 19.9 | 203.0 | 4050.0 | Male |
| 218 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | Male |
df[(df['island'] == 'Dream') & (df['body_mass_g'] > 4000)].shape
(28, 7)
df[(df['island'] == 'Dream') & (df['body_mass_g'] > 4000)].shape[0]
28
df['bill_ratio'] = df.bill_length_mm / df.bill_depth_mm
df.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | bill_ratio | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 2.090909 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 2.270115 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 2.238889 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 1.901554 |
df.isna()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | bill_ratio | |
|---|---|---|---|---|---|---|---|---|
| 0 | False | False | False | False | False | False | False | False |
| 1 | False | False | False | False | False | False | False | False |
| 2 | False | False | False | False | False | False | False | False |
| 3 | False | False | True | True | True | True | True | True |
| 4 | False | False | False | False | False | False | False | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | False | False | True | True | True | True | True | True |
| 340 | False | False | False | False | False | False | False | False |
| 341 | False | False | False | False | False | False | False | False |
| 342 | False | False | False | False | False | False | False | False |
| 343 | False | False | False | False | False | False | False | False |
344 rows × 8 columns
df.isna().any(axis=1)
0 False
1 False
2 False
3 True
4 False
...
339 True
340 False
341 False
342 False
343 False
Length: 344, dtype: bool
df_missinig = df[df.isna().any(axis=1)]
df_missinig
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | bill_ratio | |
|---|---|---|---|---|---|---|---|---|
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | NaN |
| 8 | Adelie | Torgersen | 34.1 | 18.1 | 193.0 | 3475.0 | NaN | 1.883978 |
| 9 | Adelie | Torgersen | 42.0 | 20.2 | 190.0 | 4250.0 | NaN | 2.079208 |
| 10 | Adelie | Torgersen | 37.8 | 17.1 | 186.0 | 3300.0 | NaN | 2.210526 |
| 11 | Adelie | Torgersen | 37.8 | 17.3 | 180.0 | 3700.0 | NaN | 2.184971 |
| 47 | Adelie | Dream | 37.5 | 18.9 | 179.0 | 2975.0 | NaN | 1.984127 |
| 246 | Gentoo | Biscoe | 44.5 | 14.3 | 216.0 | 4100.0 | NaN | 3.111888 |
| 286 | Gentoo | Biscoe | 46.2 | 14.4 | 214.0 | 4650.0 | NaN | 3.208333 |
| 324 | Gentoo | Biscoe | 47.3 | 13.8 | 216.0 | 4725.0 | NaN | 3.427536 |
| 336 | Gentoo | Biscoe | 44.5 | 15.7 | 217.0 | 4875.0 | NaN | 2.834395 |
| 339 | Gentoo | Biscoe | NaN | NaN | NaN | NaN | NaN | NaN |
df_clean = df.dropna()
df_clean
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | bill_ratio | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 2.090909 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 2.270115 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 2.238889 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 1.901554 |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male | 1.907767 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female | 3.445255 |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female | 3.272727 |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male | 3.210191 |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female | 3.054054 |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male | 3.099379 |
333 rows × 8 columns
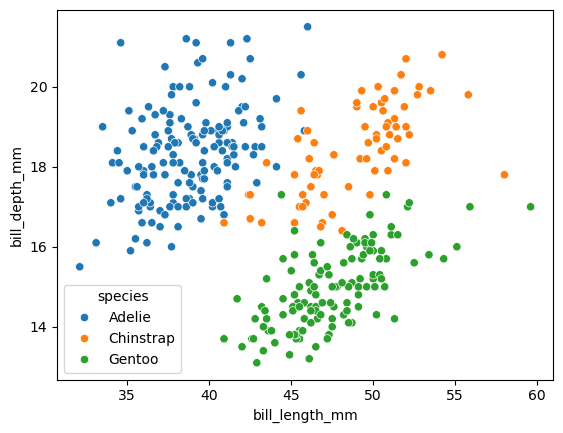
sns.scatterplot(
data = df,
x = "bill_length_mm",
y = "bill_depth_mm",
hue = "species"
)
<Axes: xlabel='bill_length_mm', ylabel='bill_depth_mm'>
import altair as alt
alt.Chart(df).mark_circle().encode(
x = alt.X("bill_length_mm",scale=alt.Scale(domain=(30, 60))),
y = alt.X("bill_depth_mm",scale=alt.Scale(domain=(12, 22))),
color = "species",
tooltip = ["species", "bill_length_mm", "bill_depth_mm", "island", "sex"]
)
/Users/Ray/opt/anaconda3/envs/math10/lib/python3.12/site-packages/altair/utils/core.py:395: FutureWarning: the convert_dtype parameter is deprecated and will be removed in a future version. Do ``ser.astype(object).apply()`` instead if you want ``convert_dtype=False``.
col = df[col_name].apply(to_list_if_array, convert_dtype=False)
/Users/Ray/opt/anaconda3/envs/math10/lib/python3.12/site-packages/altair/utils/core.py:395: FutureWarning: the convert_dtype parameter is deprecated and will be removed in a future version. Do ``ser.astype(object).apply()`` instead if you want ``convert_dtype=False``.
col = df[col_name].apply(to_list_if_array, convert_dtype=False)
/Users/Ray/opt/anaconda3/envs/math10/lib/python3.12/site-packages/altair/utils/core.py:395: FutureWarning: the convert_dtype parameter is deprecated and will be removed in a future version. Do ``ser.astype(object).apply()`` instead if you want ``convert_dtype=False``.
col = df[col_name].apply(to_list_if_array, convert_dtype=False)
brush = alt.selection_interval()
c1 = alt.Chart(df).mark_point().encode(
x = alt.X("bill_length_mm",scale=alt.Scale(domain=(30, 60))),
y = alt.X("bill_depth_mm",scale=alt.Scale(domain=(12, 22))),
color = "species:N",
tooltip = ["species", "bill_length_mm", "bill_depth_mm", "island", "sex"]
).add_params(brush)
c2 = alt.Chart(df).mark_bar().encode(
x="species:N",
y="count()"
).transform_filter(brush)
c1|c2
/Users/Ray/opt/anaconda3/envs/math10/lib/python3.12/site-packages/altair/utils/core.py:395: FutureWarning: the convert_dtype parameter is deprecated and will be removed in a future version. Do ``ser.astype(object).apply()`` instead if you want ``convert_dtype=False``.
col = df[col_name].apply(to_list_if_array, convert_dtype=False)
/Users/Ray/opt/anaconda3/envs/math10/lib/python3.12/site-packages/altair/utils/core.py:395: FutureWarning: the convert_dtype parameter is deprecated and will be removed in a future version. Do ``ser.astype(object).apply()`` instead if you want ``convert_dtype=False``.
col = df[col_name].apply(to_list_if_array, convert_dtype=False)
/Users/Ray/opt/anaconda3/envs/math10/lib/python3.12/site-packages/altair/utils/core.py:395: FutureWarning: the convert_dtype parameter is deprecated and will be removed in a future version. Do ``ser.astype(object).apply()`` instead if you want ``convert_dtype=False``.
col = df[col_name].apply(to_list_if_array, convert_dtype=False)
import plotly.express as px
px.scatter(
data_frame=df,
x = "bill_length_mm",
y = "bill_depth_mm",
color = "species",
hover_data = ["species", "bill_length_mm", "bill_depth_mm", "island"]
)
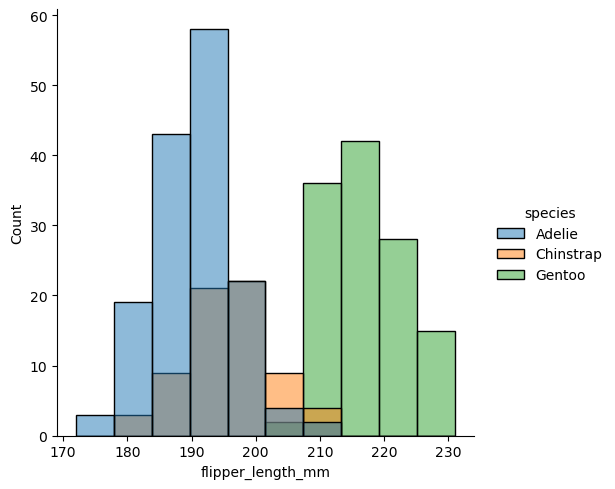
sns.displot(df, x="flipper_length_mm", hue="species")
<seaborn.axisgrid.FacetGrid at 0x18fcbfaa0>
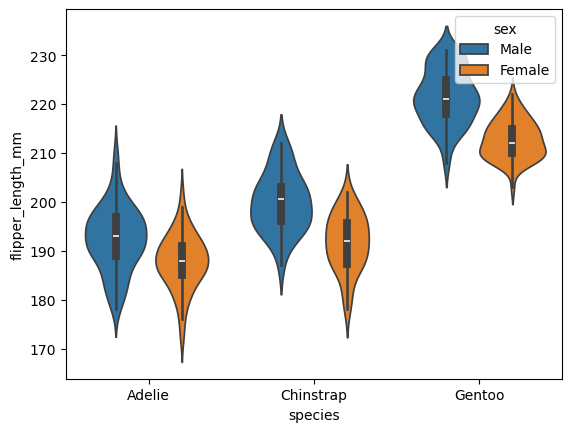
sns.violinplot(data=df, x="species", y="flipper_length_mm",hue="sex")
<Axes: xlabel='species', ylabel='flipper_length_mm'>
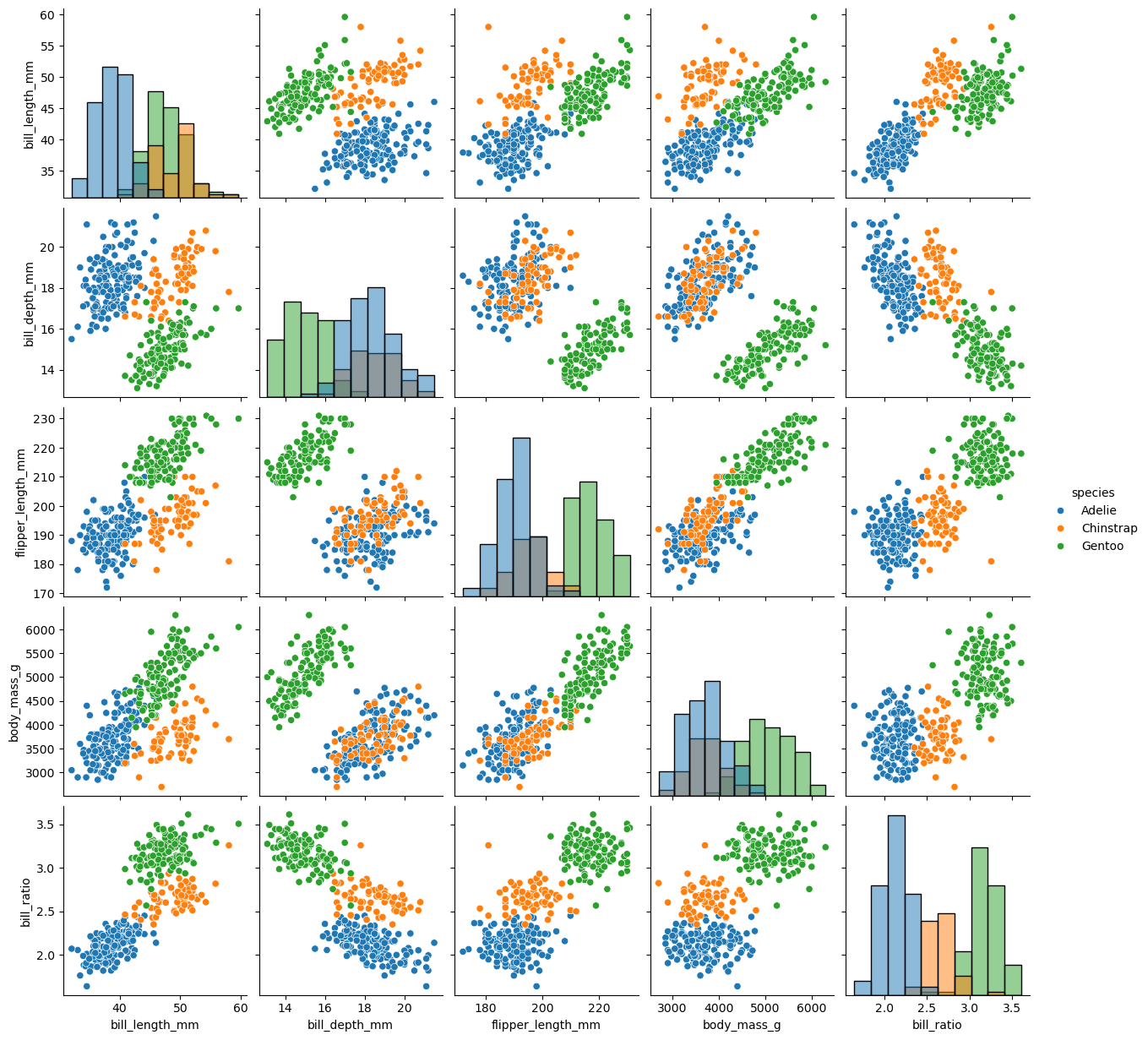
sns.pairplot(df, diag_kind="hist", hue = 'species')
<seaborn.axisgrid.PairGrid at 0x18fe09a00>